Datalad crash course

Table of content
- Goals
- Prerequisites
- Install a BIDS dataset
- Try to open a “text” file
- Saving data 💾
- Renaming a file
- Try to open a datafile and failing ❌
- Getting data
- Try to open a datafile and succeeding ✅
- Modifying data and failing ❌
- Unlocking data 🔓
- Modifying data and succeeding ✅
- Saving data again 💾
- The history of the dataset
- Pushing data and failing ❌
- Creating a remote repo on GIN 🍸
- Pushing data and succeeding ✅
- Dropping data
- Creating a dataset from scratch
- Oops! I turned my home folder into a datalad dataset… 🙈
- Oops! I accidentaly deleted some files. How can I bring them back?
- Useful tips
- Useful links
CLICK ME
... to see what I hide !!!
Goals
- install a datalad dataset and work with it
- create a datalad dataset and a remote copy of it online
The examples taken here on an MRI dataset but feel free to try this on dataset with any other type of data.
Prerequisites
In terms of technical knowledge, knowing some UNIX command line and some of the Git basics might help but are not required.
But there are things you need to install and do before the workshop:
- install datalad
- for Windows, it is preferable to use the Windows Subsystem Linux install
- if you have a Mac M1, please check this issue for a workaround in case you run into problems
- create a GIN account
- create an SSH key
- add it to your GIN account:
- basic configuration
Pre-flight checks
Checks to make sure everything is set up correctly
In a terminal, make sure that you have a version of datalad >= 0.13
datalad --version
Example output
datalad 0.13.4
Try to install a dataset from GIN
datalad install -s git@gin.g-node.org:/cpp-lln-lab/CPP_visMotion-raw.git \
~/CPP_visMotion-raw
The first time you do this, it will ask you some confirmation about using your SSH key to connect to GIN. This is normal and you can safely say “yes”.
Example output
install(ok): /home/remi/CPP_visMotion-raw (dataset)
If this install works, you can remove the dataset with.
rm -rf ~/CPP_visMotion-raw
Install a BIDS dataset
Choose a BIDS dataset you want to install.
From GIN 🍸
- a public fMRI dataset from our lab
- your own dataset from GIN: it should be in a private repository you should have access to in the CPP LLN lab organization on GIN
To install a dataset from GIN make sure you copy the SSH url:

From openneuro
- a dataset from openneuro
- Each dataset has a “download” link that will give you the datalad command to install it.
- See for example this dataset
Install it
This is done with the datalad install command.
In its simplest form it just requires a source URL to install a dataset from.
datalad install -s ${url}
What's with the dollar $ and the curly braces {}?
This is how you can call previously stored variables in bash (the language used by default in most terminals).
url="git@gin.g-node.org:/cpp-lln-lab/CPP_visMotion-raw.git"
datalad install -s ${url}
So if the URL you copied is
git@gin.g-node.org:/cpp-lln-lab/CPP_visMotion-raw.git, then you just need to
type:
Example
datalad install -s git@gin.g-node.org:/cpp-lln-lab/CPP_visMotion-raw.git
You can also specify the folder where to install the dataset.
datalad install -s ${url} \
${where_to_install}
Example
datalad install -s git@gin.g-node.org:/cpp-lln-lab/CPP_visMotion-raw.git \
~/gin/CPP_visMotion-raw
If everything went smoothly the folder structure and the files should now be on your computer and you can browse them in your file explorer or via your terminal.
Try to open a “text” file
If we have a quick look at the content of our data, we can see that it contains some plain text file like the README, or some JSON or TSV.
Example
cd ~/gin/CPP_visMotion-raw
tree -L 1
Example output
.
├── CHANGES
├── dataset_description.json
├── participants.json
├── participants.tsv
├── README
├── sub-con07
├── sub-con08
├── sub-con15
└── task-visMotion_bold.json
Try to open and edit one of these text files with your favorite text editor: for example adding some details in the README (there are rarely enough details in those anyway).
You see you can open, modify and save that file.
Saving data 💾
Now let’s see what datalad says about the status of our dataset.
datalad status
Example output
modified: README (file)
When you want to save the changes you made to a dataset, simply use the
datalad save command.
You should also add a specific message to your save, to describe the content of the save.
Here are some tips on how to write good save messages.
Note that “saves” are often referred to as “commits” and that are thus accompanied by a “commit message”.
Example
datalad save -m 'Update README'
Example output
add(ok): README (file)
save(ok): . (dataset)
action summary:
add (ok: 1)
save (ok: 1)
How often should I save? What should I save?
Note that you can save specific files only with datalad
datalad save -m ${your_message} ${path_to_file_or_folder_to_save}
An advice that you might hear is:
Commit early. Commit often. A version control advice, not a relationship advice.More serious advices can be found in the Turing way book
For the GIT users out there...
Datalad does not have what is called a "staging area" in GIT. So there is no need to "add" a file before it can be committed. More info on this here.
Even though you CAN use the typical holy trinity of git commands (git
add / git commit / git push) when working
within a Datalad dataset, I would recommended using typical Datalad command like
datalad save, unless you are sure of what you are doing.
However it should be completely fine to use Datalad commands within a pure git repository.
Renaming a file
Now let’s rename one of our non-text file and see how that goes.
This can be done via your usual file explorer or via the terminal with the move
command (mv).
mv -v sub-con07/ses-01/anat/sub-con07_ses-01_T1w.nii sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii
Example output
renamed 'sub-con07/ses-01/anat/sub-con07_ses-01_T1w.nii' -> 'sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii'
OK that seems to work. So let’s save that change too.
datalad save -m 'Rename anat subject con07'
Example output
delete(ok): sub-con07/ses-01/anat/sub-con07_ses-01_T1w.nii (file)
add(ok): sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii (file)
save(ok): . (dataset)
action summary:
add (ok: 1)
delete (ok: 1)
save (ok: 1)
Try to open a datafile and failing ❌
Let’s try to open a file that is not “just” a text file.
In this example we could try to open a nifti image (.nii) with MRIcron,
fsleyes, SPM CheckReg…
If you have installed an EEG dataset you can try to load it with the tools you usually use to quickly view those files.
Apparently fsleyes is not happy when trying to open the T1w file of an installed dataset.
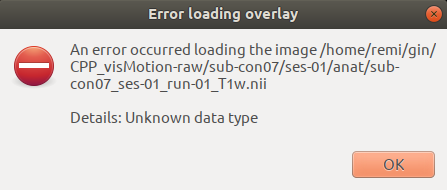
🚨 Under the hood: remote content 🚨
This is because even though, the file shows up on your computer its “content” has not been downloaded on your computer.
As one of our lab member once said:
datalad installgives you bunch of “ghost fileS” 👻🗄️ that have the right shape but no substance.
This is also why installing the whole dataset was so quick: very little data was actually downloaded.
To fix this issue, we must get the content from where the dataset was installed.
Getting data
To get the content of a file you use the datalad get command that must be run
from within the dataset (from a folder in the dataset).
datalad get ${path_to_the_folder_or_file}
Example
datalad get sub-con07/anat/*T1w.nii
What's with the star *?
That's a bash thing to allow you to say "any sequence of characters". So in our case that would mean any file in the
anat folder that ends in
T1w.nii.
More on bash "globbing"
here
Example output
Total: 0%| | 0.00/25.2M [00:00<?, ? Bytes/s]
Get sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii: 34%|████████ | 8.58M/25.2M [00:00<00:00, 64.9M Bytes/s]
And eventually:
get(ok): sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii (file) [from origin...]
Note
Depending on the color highlighting of your terminal, you might even see a difference on the color of the file whose content you just got.
Try to open a datafile and succeeding ✅
You can now browse the actual content of that file.
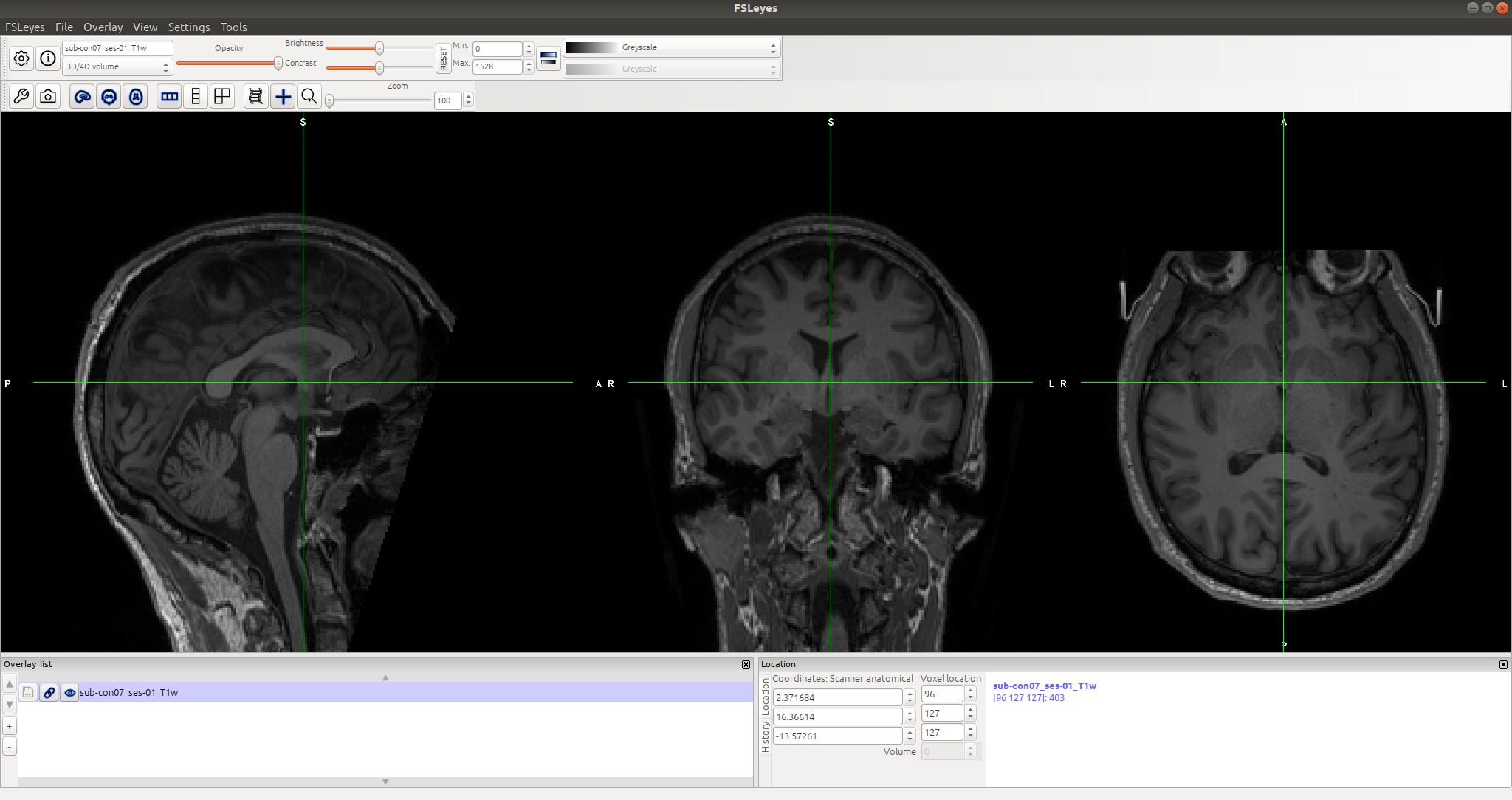
Modifying data and failing ❌
Let’s now try to something basic on this image to modify it, like reorienting it with SPM by setting the origin of the image to the anterior commissure.
In SPM, you can open the image with the CheckReg tool, locate the anterior
commissure and then right click Reorient --> Set origin to cross-hair and then
select the images to apply this change to.
SPM will try to modify the header of the image to implement the requested change and will fail majestically in the process.
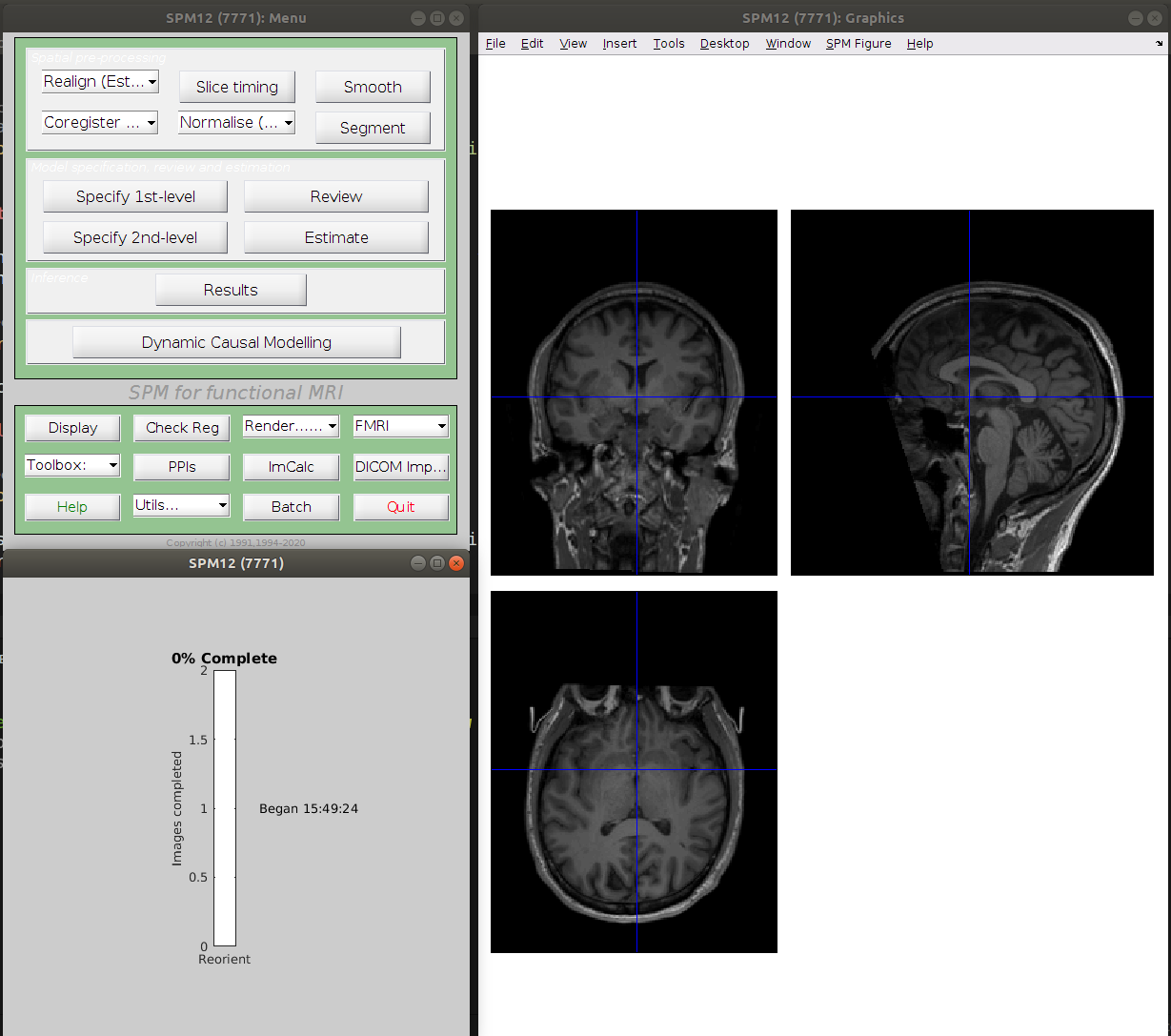
------------------------------------------------------------------------
14-Feb-2022 15:49:23 - Running job #1
------------------------------------------------------------------------
14-Feb-2022 15:49:23 - Running 'Reorient Images'
Error: Permission denied
There was a problem writing to the header of
"/home/remi/gin/CPP_visMotion-raw/sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii"
14-Feb-2022 15:49:24 - Failed 'Reorient Images'
Error using nifti/create (line 26)
Unable to write header for "/home/remi/gin/CPP_visMotion-raw/sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii".
In file "/home/remi/matlab/SPM/spm12/@nifti/create.m" (v7147), function "create" at line 26.
In file "/home/remi/matlab/SPM/spm12/spm_get_space.m" (v6379), function "spm_get_space" at line 51.
In file "/home/remi/matlab/SPM/spm12/config/spm_run_reorient.m" (v6078), function "spm_run_reorient" at line 33.
The following modules did not run:
Failed: Reorient Images
Error using MATLABbatch system
Job execution failed. The full log of this run can be found in MATLAB command window, starting with the lines (look for the line
showing the exact #job as displayed in this error message)
------------------
Running job #1
------------------
Error while evaluating Menu Callback.
The important error in there is Error: Permission denied.
However if we check the file permissions with our file explorer / finder (or
with the terminal with the list command ls -l), we can see that we have “write
access” to all of these files. And yet we can edit the simple “text” file and
not the non “text” data.
Example
ls -l sub-con07/ses-01/anat
Example output
-rwxrwxr-x 1 remi remi 2170 Feb 14 14:23 sub-con07_ses-01_T1w.json
lrwxrwxrwx 1 remi remi 139 Feb 14 14:23 sub-con07_ses-01_run-01_T1w.nii -> ../../../.git/annex/objects/j5/G3/MD5E-s25166176--d5dec5aad67f659c2f218f096cb4b8d4.nii/MD5E-s25166176--d5dec5aad67f659c2f218f096cb4b8d4.nii
The rwxrwxrwx at the beginning of these lines defines if we have read (r),
write (w), execute (x) rights on those files. More on unix file permission
here.
🚨 Under the hood: annexed files 🚨
We cannot edit this file, because its content has been “annexed”.
You can usually tell a file is annexed when listing it in a terminal with
ls -l or when viewing it with tree.
lrwxrwxrwx 1 remi remi 139 Feb 14 14:23 sub-con07_ses-01_run-01_T1w.nii -> ../../../.git/annex/objects/j5/G3/MD5E-s25166176--d5dec5aad67f659c2f218f096cb4b8d4.nii/MD5E-s25166176--d5dec5aad67f659c2f218f096cb4b8d4.nii
- the
lmeans that the file you are looking at is a “link” - the
->at the end shows what this links to: in this case a file in the.git/annexin the root of the dataset - the file size (the 5th column on the
lsoutput above) is usually a hint that the content of that file has been annexed: in this case the “size” of this nifti image is less than that a very simple JSON file (which sounds unlikely if the content were not annexed).
A hand wavy explanation of this annexation behavior is that Datalad in fact acts a bit as as “a wrapper” around:
gitthat is good for version controlling text based files (like code, TSV, JSON, …) but start struggling with non-text files and large number of filesgit-annexthat helps for the version control of data files.
To be able to edit this file, we must first unlock it.
Unlocking data 🔓
This is done with the datalad unlock command.
datalad unlock ${path_to_the_folder_or_file}
So in our case:
Example
datalad unlock sub-con07/ses-01/anat/*T1w.nii
Example output
unlock(ok): sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii (file)
The result of unlocking the file will be visible in different ways.
On a tree or a ls command, it won’t show as a link but as an “actual” file.
Example
tree -L 3 sub-con07
Example output
sub-con07
└── ses-01
├── anat
│ ├── sub-con07_ses-01_T1w.json
│ └── sub-con07_ses-01_run-01_T1w.nii
└── func
├── sub-con07_ses-01_task-visMotion_bold.nii -> ../../../.git/annex/objects/mJ/PF/MD5E-s370137952--89a6190d568fa6cbb0fd1f23eb763b0c.nii/MD5E-s370137952--89a6190d568fa6cbb0fd1f23eb763b0c.nii
└── sub-con07_ses-01_task-visMotion_events.tsv
Also an ls -l would now show you the actual actual size of this file.
If you use the datalad status command, datalad will tell that this file has
been modified because it was taken out of the annex.
Example output
modified: sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii (file)
Modifying data and succeeding ✅
Now let’s try to modify this file again.
This time SPM seems happy.
------------------------------------------------------------------------
14-Feb-2022 21:17:17 - Running job #1
------------------------------------------------------------------------
14-Feb-2022 21:17:17 - Running 'Reorient Images'
14-Feb-2022 21:17:18 - Done 'Reorient Images'
14-Feb-2022 21:17:18 - Done
Now let’s see what datalad says about the status of our dataset.
datalad status
Example output
untracked: sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w_reorient.mat (file)
modified: sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii (file)
Now datalad tells us about the files that have been modified and also about the new file created by SPM.
Saving data again 💾
Example
datalad save -m 'setting origin to anterior commissure for subject 07'
Example output
add(ok): sub-con07/ses-01/anat/sub-con07_ses-01_T1w_reorient.mat (file)
add(ok): sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii (file)
save(ok): . (dataset)
action summary:
add (ok: 2)
save (ok: 1)
Note that saving a file, relocks it in the annex.
tree -L 3 sub-con07/anat
Example output
sub-con07/ses-01/anat
├── sub-con07_ses-01_T1w.json
├── sub-con07_ses-01_run-01_T1w.nii -> ../../../.git/annex/objects/VK/WP/MD5E-s25166176--f4db32e160f289d5cc01d3a23aadf4d0.nii/MD5E-s25166176--f4db32e160f289d5cc01d3a23aadf4d0.nii
└── sub-con07_ses-01_T1w_reorient.mat -> ../../../.git/annex/objects/68/79/MD5E-s312--85774df306f765822b4c203bca41bb13.mat/MD5E-s312--85774df306f765822b4c203bca41bb13.mat
See also how the new .mat file was annexed as well.
The history of the dataset
You can check all the past changes that have been made to the dataset with the
git log command. Type q to exit this “log mode”.
If you want a less verbose version of the log: git log --oneline.
097059d (HEAD -> master) setting origin to anterior commissure for subject 07
c4a039e Rename anat subject con07
8c82a0f Update README
b003e21 (origin/master, origin/HEAD) initial save
2a72d36 Instruct annex to add text files to Git
52c0844 [DATALAD] new dataset
Given that more advanced usage of datalad allow you to “travel in time” 🕥🔄 through the history of a dataset to see what it looked like at a given commit, you can quickly imagine that having good commit messages helps you knowing what is your “best destination”.

Pushing data and failing ❌
Now that we have made a change we want to back it up online to make sure it is not lost in case our computer catches on fire. 💻🔥
datalad push --to ${sibling_name}
When you install a dataset from somewhere, the dataset that lives on your
computer is usually called the local copy (when you talk to another human being)
or the local clone (when you talk to a git user) or a local
sibling
(when you talk to a datalad user). The sibling you installed from is usually
called a remote sibling and often nicknamed origin.
So to update the origin with our changes we would do:
Example
datalad push --to origin
Odds are that, unless you are the owner of the origin sibling, you encountered
an error that prevented from updating the online dataset.
🚨 Under the hood: siblings 🚨
Very often you won’t have the write access to make changes to the repository where you got a dataset from.
If however you wanted to keep track of your own changes, you can have your own copy of that dataset on GIN by creating a sibling repository on GIN.
The good news is that unless the data owner has given you the right to do so, it is extremely unlikely you will break or destroy someone else’s data.
Creating a remote repo on GIN 🍸
So let’s create a repository on GIN.
To make your life easier do not initialize any file in this repository.
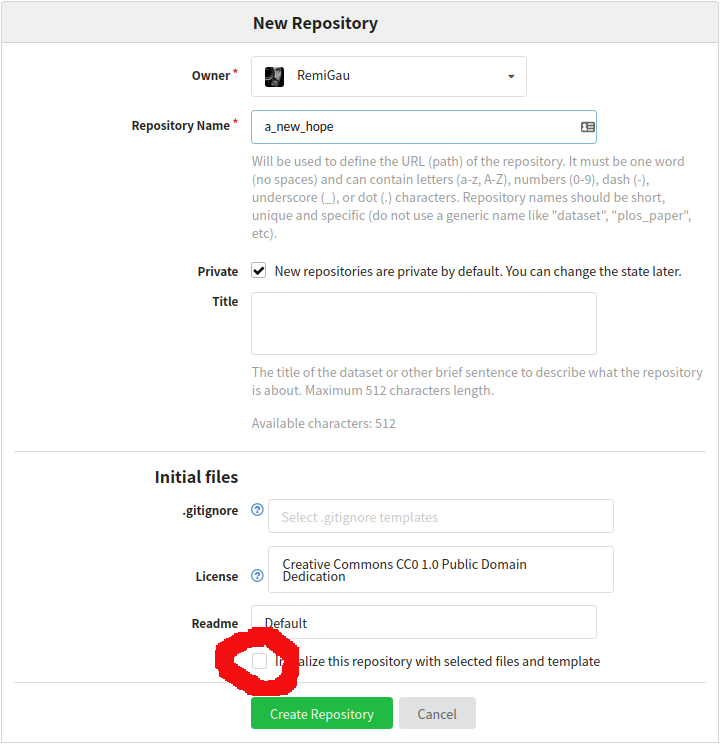
You must then copy the SSH URL.
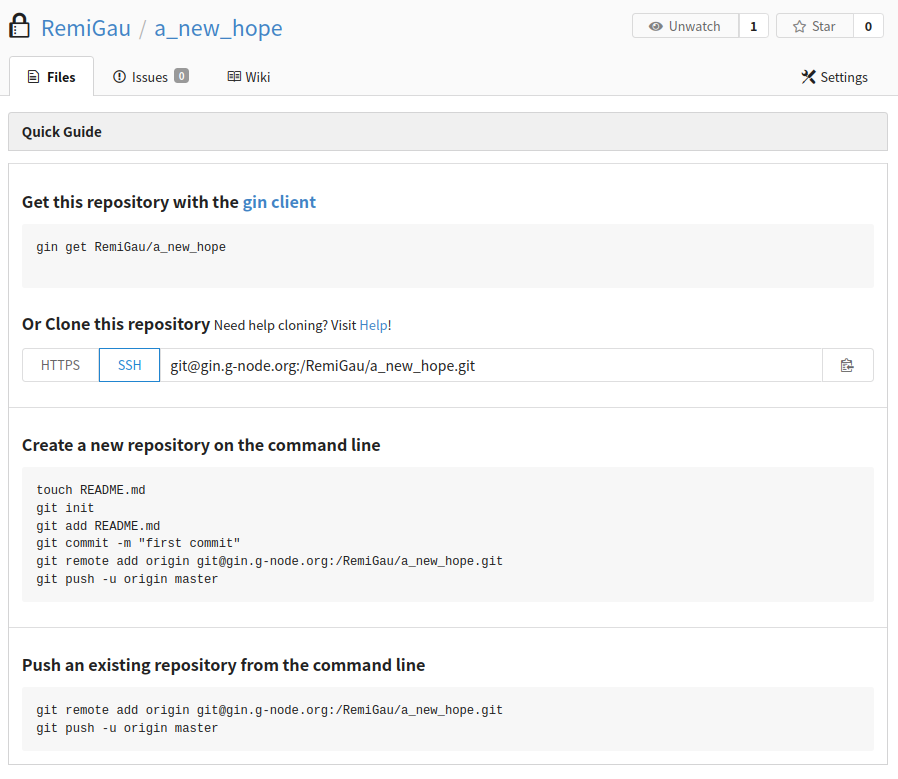
You then need to let your local sibling that there is another repository. This
is done with the datalad siblings add command.
datalad siblings add --name ${name} --url ${url}
Example
datalad siblings add --name a_new_hope --url git@gin.g-node.org:/RemiGau/a_new_hope.git
Example output
[INFO ] Could not enable annex remote a_new_hope. This is expected if a_new_hope is a pure Git remote, or happens if it is not accessible.
[WARNING] Could not detect whether a_new_hope carries an annex. If a_new_hope is a pure Git remote, this is expected.
.: a_new_hope(-) [git@gin.g-node.org:/RemiGau/a_new_hope.git (git)]
Pushing data and succeeding ✅
You should now be able to push your changes to this new sibling.
datalad push --to a_new_hope
Example output
Transfer data to 'a_new_hope': 50%|██████████████████████▌ | 2.00/4.00 [00:00<00:00, 8.36k Steps/s]
Total: 0%| | 0.00/1.17G [00:00<?, ? Bytes/s]
Copy sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii: 54%|██████████████▏ | 13.7M/25.2M [00:09<00:07, 1.62M Bytes/s]
And eventually:
copy(ok): sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii (file) [to a_new_hope...]
copy(ok): sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w_reorient.mat (file) [to a_new_hope...]
publish(ok): . (dataset) [refs/heads/git-annex->a_new_hope:refs/heads/git-annex e2c7f02..0a33ba9]
publish(ok): . (dataset) [refs/heads/master->a_new_hope:refs/heads/master [new branch]]
action summary:
copy (ok: 2)
publish (ok: 2)
If you now refresh the web page of your GIN repository, it should display the content of your data set.
My repo in GIN looks nothing like what I have on my computer!!
This might be because you are actually viewing the annexed content of your dataset that lives on the "git-annex branch": click here to see how to fix it (the fix relates to GitHub repositories but works on GIN too).
Note however that your remote siblings contains the content for the data you had in your local sibling.
You can use the git annex list to know where you have copy of what
Example
git annex list .
Example output
here
|a_new_hope
||origin
|||web
||||bittorrent
|||||
XXX__ sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii
XX___ sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w_reorient.mat
__X__ sub-con07/ses-01/func/sub-con07_ses-01_task-visMotion_bold.nii
__X__ sub-con08/ses-01/anat/sub-con08_ses-01_T1w.nii
__X__ sub-con08/ses-01/func/sub-con08_ses-01_task-visMotion_bold.nii
__X__ sub-con15/ses-01/anat/sub-con15_ses-01_T1w.nii
__X__ sub-con15/ses-01/func/sub-con15_ses-01_task-visMotion_bold.nii
So if you wanted to make an entire copy of the original dataset you would to
get that content first from the origin and then push it to the new one.
datalad get .
datalad push --to a_new_hope
We are not going to do that now, otherwise we are going to be at this until the cows come home. 🐮 ➡️ 🏠
Dropping data
Sometimes our dataset get big and we usually start transferring them (or part of them) on external hard drives and then we start having to keep track of which file is backed up where, when was the back up done.
Then we get told we need several back ups in different locations, so now we have to track several copies of the same dataset and make sure that they are all in synch with each other.
Datalad allows you to keep a local copy of your data and to keep your different back up (as siblings) synched. But if you need to make some space on your computer without having to delete a dataset, Datalad also allows you to drop the content of files locally.
Conceptually this is a bit like the opposite of the datalad get command we saw
earlier that would get the content of file from a sibling.
datalad drop will instead remove the content of a file from the annex provided
that this content exists in another sibling.
To know how much data exists in your annex currently you can use
datalad status --annex all.
Example output
7 annex'd files (24.0 MB/1.1 GB present/total size)
nothing to save, working tree clean
You can then drop the content of specific files.
datalad drop ${path_to_file}
The nuclear option 💥☢️ is to drop the entire dataset.
Example
datalad drop .
The . is NOT a typo but means “this folder”. A bit like .. means the parent
folder of where we are now.
Example output
drop(ok): /home/remi/gin/CPP_visMotion-raw/sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w.nii (file) [locking a_new_hope...]
drop(ok): /home/remi/gin/CPP_visMotion-raw/sub-con07/ses-01/anat/sub-con07_ses-01_run-01_T1w_reorient.mat (file) [locking a_new_hope...]
action summary:
drop (notneeded: 20, ok: 2)
Creating a dataset from scratch
OK that’s cool and all to be a data consumer and get data from somewhere else and modify it. But during my PhD I will acquire new data. So how do I start a Datalad dataset from scratch?
To do that you must first create an empty dataset to provide a home for your
data. This can be done with the datalad create command.
datalad create ${name_of_the_dataset}
This will create a new folder and initialize a Datalad dataset in it.
What if I already have a folder with my data and I want to create a dataset in there?
For example
cd ~/gin/raw
tree -d
Example output
.
└── sub-001
├── ses-01
│ ├── anat
│ └── func
└── ses-02
└── func
In this case…

datalad create --force .
Example output
[INFO ] Creating a new annex repo at /home/remi/gin/raw
create(ok): /home/remi/gin/raw (dataset)
Backing it up online
The following steps are things that we have seen in the previous steps. So that’s easy peasy, lemon squeezy 🗜️🍋 OK… sort of lemon squeezy… I very often have to look those up to make sure I don’t mess anything.
datalad save -m 'save all my data'- Create an online repository on GIN
datalad siblings add --name origin --url ${url}datalad push --to origin
Grab a coffee and look at that progress bar go! 🚀
Oops! I turned my home folder into a datalad dataset… 🙈
It happened so quickly!
#TrueStory
🚨 DON’T TRY THIS AT $HOME 🚨 (pun intended)
# when not given any argument
# the "change directory" command will bring you to your home folder
cd
# if you now force create a datalad dataset,
# you are basically telling datalad to version control your whole home folder
datalad create --force .
Imagine you typed the 2 commands above. Now datalad wants to track all the files in your computer.
FYI: if by any chance you were planning to use datalad to back up you computer, there are better tools for this.
More seriously. How to fix this?
OK that gives us the occasion to do a shallow dive into the plumbings of Datalad. 🕵️♀️
🚨 Under the hood: the .git folder 🚨
Basically all the history of your Datalad dataset is store in the hidden folder
.git that lives in the root of your dataset.
Additionally, data that has been annexed AND locked is stored in .git/annex.
Note that you might need to change some settings in your file explorer / finder
to be able to see hidden folders, in a terminal you can see those folders by
using ls -a).
So if you have created a Datalad dataset in the wrong place, you can simply
delete the .git and .datalad folders. HOWEVER if you have already saved
some data in that dataset, make sure you unlock it before removing the folder
otherwise you will lose the data.
If you want an hour long deep dive on the content of the .git folder,
check this video.
Oops! I accidentaly deleted some files. How can I bring them back?
If you deleted some files but have not yet saved those changes, there is an easy way to bring them back.
In case you don’t remember type git status and it should show a listing of
file changes including deletion, like the example below.
Example output
On branch master
Your branch is up to date with 'origin/master'.
Changes not staged for commit:
(use "git add/rm <file>..." to update what will be committed)
(use "git restore <file>..." to discard changes in working directory)
modified: README.md
deleted: images/use_the_force.jpg
no changes added to commit (use "git add" and/or "git commit -a")
A lot of git commands give you hint on what to type to do certain things.
Here it tells you that undo this deletion I would have to type:
git restore images/use_the_force.jpg
Useful tips
If you don’t remember the specifics of a command, type the name of that command
followed by --help. For example datalad create --help.
More info on how to get help.
Useful links
- Datalad handbook
- Read at least the 2 first chapters. This will save from a lot of pain and confusion.
- Datalad cheat sheet
- Datalad research data management course
- Basic file system operations with Datalad
- Datalad aliases - unofficial
